Calcium imaging is only an indirect readout of neuronal activity via fluorescence signals. To estimate the true underlying firing rates of these neurons, methods for “spike inference” have been developed. They are useful to denoise calcium imaging data and make them more interpretable. A few years ago, I developed CASCADE, a supervised method for spike inference based on deep networks. I have been been updating and maintaining CASCADE ever since, and this maintenance work has been a starting point for several collaborations and friendly interactions during the last years (for example, check out this recent preprint on spike inference from spinal cord neurons).

Originally, the CASCADE algorithm was trained on a ground truth database that consisted primarily of recordings with GCaMP6. However, how will the algorithm perform on new indicators like GCaMP8, with its much faster rise times? To address this question, I now trained CASCADE models on GCaMP8 ground truth and evaluated whether these models performed better than previous models. The short answer is – the retrained models performed clearly better:
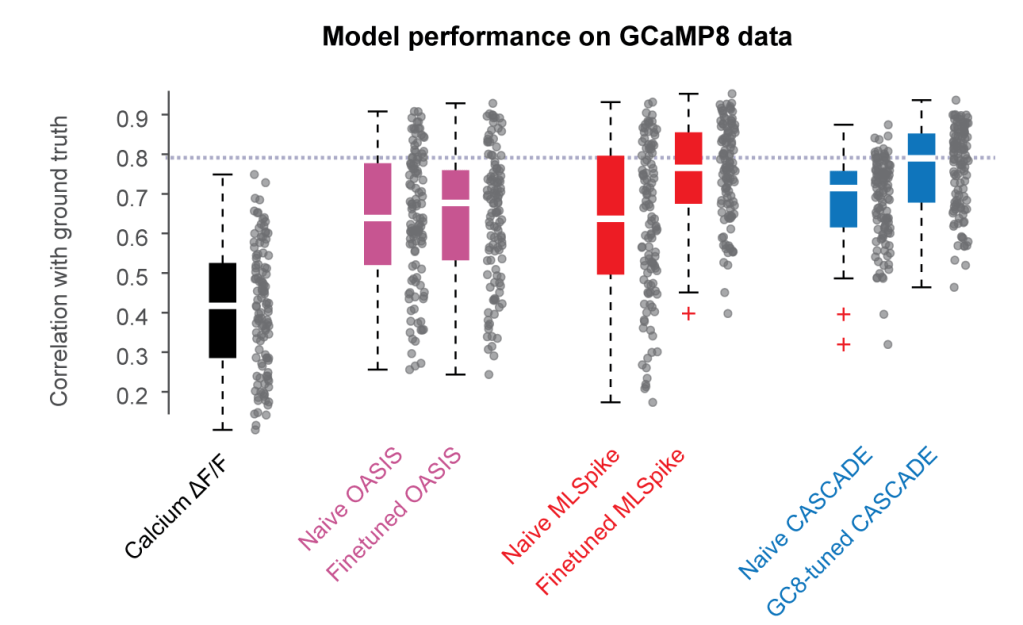
I’m currently in the process of dissecting this improvement: Is it due to differences in rise times, different non-linearities or other differing properties of the two indicator families? The results of these analyses turned out to be more fascinating than I expected and therefore take more time to understand and analyze, but I’m planning to have this analysis written up within the next 3-6 months.
In the meantime, however, feel already free to use the new CASCADE models specifically trained with and for GCaMP8 – they really make predictions better! (And please only apply these models to GCaMP8 data; the previous models are still better for anything with GCaMP6!)
You will find the new models for CASCADE as usual in the list of available pretrained models. For example, instead of the GCaMP6-trained model Global_EXC_30Hz_smoothing25ms, indicate the GCaMP8-trained model GC8_EXC_30Hz_smoothing25ms_high_noise to infer spike rates with the predict() function of CASCADE.
A technical note: These models are pretrained on all available GCaMP8-ground truth, mixing together GCaMP8f, GCaMP8m and GCaMP8s. This procedure results in more robust models due to the larger ground truth database, but absolute inferred spike rates are slightly biased due to the different spike-event fluorescence amplitudes for the three indicators (-30% underestimate for GCaMP8f and GCaMP8m, +60% overestimate for GCaMP8s). In the near future, CASCADE will also include models specific for each of those indicators. These models will probably be slightly less robust but will provide less biased spike rates. However, if you are not specifically interested in very precise absolute spike rates, I would for now recommend the general GCaMP8 models that are already available. They are not only robust and very powerful but also provide a good rough estimate of absolute spike rates.
Update 2024-09-19: Models pretrained with specific indicators (e.g., GC8s_EXC_30Hz_smoothing25ms_high_noise for GCaMP8s) are now available online. Check out the full list of models via the Cascade code or via this continuously updated list on Github.
Update 2025-03-11: A preprint describing the analyses, the pretrained models and their applications to GCaMP8 is now on bioRixv: https://www.biorxiv.org/content/10.1101/2025.03.03.641129.
If you have questions, please reach out via comments on the blog, issues on Github or via email!
Nice! Where would GCaMP6s be in figure 2 in terms of correlation with ground truth?
But to give you some numbers (with the caveat of interpretation that I mentioned before): The median correlation with ground truth (± s.d. across neurons), for this specific level of shot noise:
GCaMP8f with CG8-trained CASCADE: 0.72±0.10
GCaMP8m with CG8-trained CASCADE: 0.79±0.12
GCaMP8s with CG8-trained CASCADE: 0.83±0.07
GCaMP6s (Huang et al.) with GC6-trained CASCADE: 0.68±0.11
GCaMP6s (Chen et al.) with GC6-trained CASCADE: 0.59±0.08
These statistics are now properly quantified and reported in https://www.biorxiv.org/content/10.1101/2025.03.03.641129v1 in Fig. 1 Suppl. 6.
Hi – that’s a good question, but there is no good answer. These analyses above are based on GCaMP8-based ground truth recorded by a single experimenter (Marton Rozsa). There are ground truth datasets for GCaMP6s recorded by other experimenters in the same brain area (one dataset from the original GCaMP6 paper, another dataset from the Allen Institute), for which I could run the same analysis, but it is difficult to compare these datasets. For example, experimenter 2 could have applied more sensory stimuli, thereby increasing the overall activity level and making it easier to achieve high correlation values. Or they could have been more conservative, discarding recordings with significant motion artifacts – that’s always a very arbitrary and subjective decision. Therefore, I’m very hesitant to make any quantitative comparisons of indicator quality based on these values.